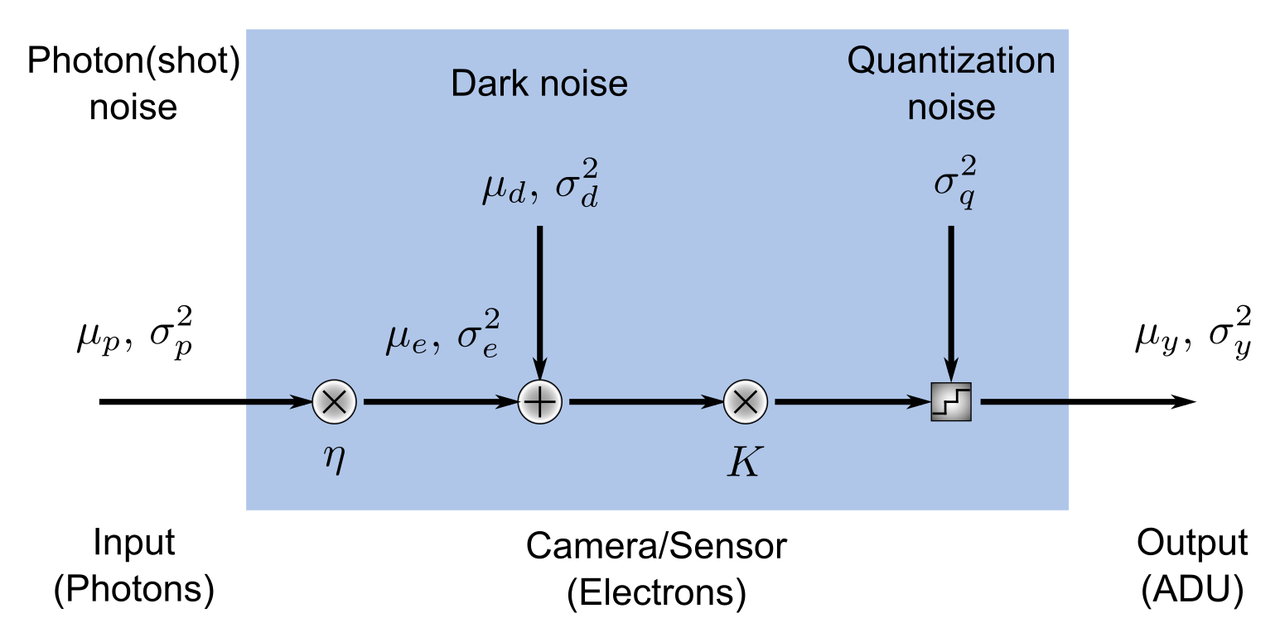
Accessing the Raspberry Pi camera image sensor
WARNING You can easily ruin your Raspberry Pi camera module by following the steps in this post. Proceed with caution.
Over the past half year I have been making slow but steady progress on my lensless imager project. The purpose, aside from having a bit of fun, is to create an imaging system for basic cell biology that doesn't use an expensive microscope objective.
A lensless imager works just as its name implies: an image sensor records the scattered light from a microscopic, transparent object and computationally reconstructs an image of that object, all without a lens. The best resolutions are achieved when the object is relatively close to the image sensor. Ideally, the separation between the object and the sensor's pixels would be at most about one millimeter. This limit is partly determined by the light source's spatial coherence, but also by the fact that high resolution is achieved by recording the scattered light at very large angles, which is possible only when the sample is close to the sensor.
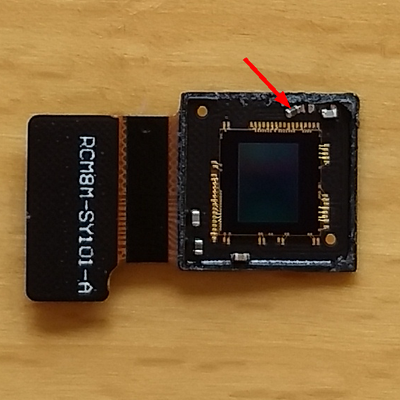
Today I had a bit of free time so I decided to see whether I could remove the housing that surrounds the Raspberry Pi camera's sensor. The Raspberry Pi Camera Module version 2 sensor is a Sony IMX219 color chip. Directly above the sensor is a filter, a lens, and the housing for both of these that prevent me from placing anything closer than about half a centimeter from the sensor plane. If I would want to use this camera for the lensless imager, then the housing would have to go.
Now, even without the housing the Raspberry Pi camera is not necessarily the best option for the project because the IMX219 is a color sensor. This means that there is a Bayer filter over its pixels, which would cut the resolution of the imager since I would be using a very narrow band light source. Effectively, only a quarter of the pixels would be receiving any light. Regardless, I had a spare second camera and it interfaces well with the Raspberry Pi, so I figured it would make for a good prototype.
As you will see, I did slightly damage my sensor, though it seems to still work. You can easily ruin your camera module or cut your finger by following these steps, so proceed at your own risk.
Step 0: The Raspberry Pi Camera Module V2
In the picture below you see my Raspberry Pi Camera Module. From above, you can see the small circular aperture with the lens immediately behind it. The sensor is inside the small gray rectangular housing that is attached to the control board by a small ribbon cable (to its right) and a bit of two-sided sticky foam tape (underneath the sensor; not visible).
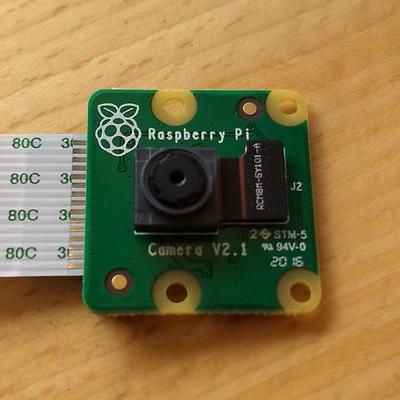
Step 1: Remove the main ribbon cable
To make working on the board a bit easier, I removed the white ribbon cable that attaches the module to the Pi. I did this by pulling on the black tabs on the two ends of the connecter until the cable is easily removed. I labeled the sides of the ribbon cable just in case.

Step 2: Detach the sensor's ribbon cable
Next, I used my finger and thumb nail to remove the small ribbon cable that attaches the sensor to the control board. I essentially applied a small torque to the bottom edge of the connector until it just "popped" up, as seen in the second image below.
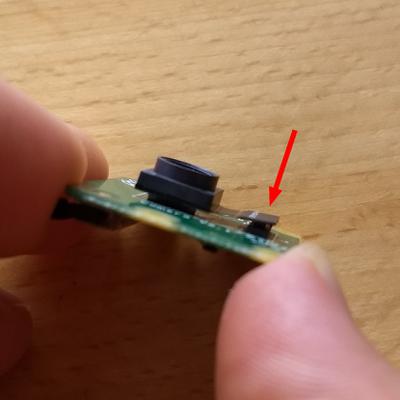
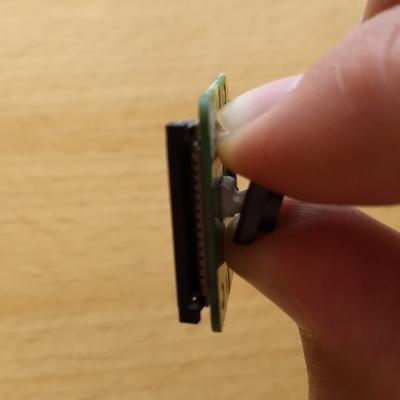
Step 3: Remove the sensor from the control board
In the third step, I used my thumbnail to gently pry the sensor from the control board. The sensor is attached with some two-sided sticky tape and may need a few minutes of work to come free.
Step 4: Remove the rectangular housing
In this step you risk cutting your finger, so please be careful.
The housing around the sensor is glued. To remove it, you will need to gently work a knife (or, better yet, a thin screw driver) between the housing and the sensor board, taking care not to let the blade go too far into the housing and possibly ruining one of the resistors or wire bonds.

Once you get a knife between the two, try popping the housing off of the sensor.

When I did this I cut on three sides of the housing, but in retrospect I should have only cut on the side opposite the ribbon cable and pried the other sides loose. This is because I damaged a small resistor when the knife blade went too far into the housing. You can see this below and, at the same time, get an idea of the layout of the sensor board so you know where you can and can't cut.
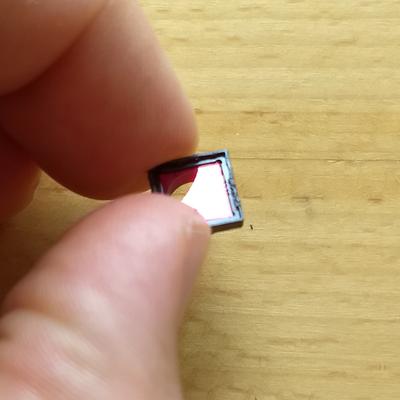
If you have the normal version of the camera, then you can also find the IR blocking filter inside the housing.
Fortunately for me the camera still works, despite the damaged resistor. I can now place samples directly on the sensor if I wanted to, though the wire bonds from the sensor to its control board appear quite fragile. For this reason, it may make more sense to build a slide holder that holds a sample just above the surface without touching it. For now, I can use this exposed sensor to prototype different methods for mounting the sample.